Narragansett Bay Adult Oyster DNA Extractions Part 1
DNA Extraction of Adult Oyster Tissue - Part 1
~10 wild adult oysters were collected from 5 populations in Narragansett Bay
- Narrow River
- Green Hill Pond
- Barrington River
- Kickemuit
- Mary C. Donovan Marsh
Mantle and gill tissue were dissected from the oysters. Dissection protocol can be accessed here.
DNA Extractions
Completed on January 18, 2021
Zymo Research Quick-DNA Miniprep Plus used for DNA extractions of adult oyster tissue
- Pull samples out of -80 freezer and put on ice
- NAR_7 M2
- BAR_5 M2
- BAR_7 M2
- GHP_8 M2
- KIC_6 M1
- Pull Proteinase k out of upright -20 freezer in Puritz Zymo Reagents box and put on ice
- Pull out Blue Solid Tissue Buffer from Zymo Research Quick DNA Miniprep kit
- To each original tube with the tissue piece, add 95 ul of nuclease-free water, 95 ul of solid tissue buffer, and 10 ul of Proteinase k
- Vortex for 10 seconds and spin down on mini centrifuge
- Place tubes in thermomixer at 55 deg C shaking at 1200 rpm
- Check tissue after 30 minutes
- The liquid was pretty cloudy, so I added another 15 minutes and then another 30 minutes
- After 1 hour and 15 minutes, the liquid still looked cloudy so I decided to move forward with the dissection protocol
- Check tissue after 30 minutes
NAR_7 M2:  BAR_5 M2:
BAR_5 M2:  BAR_7 M2:
BAR_7 M2:  GHP_8 M2:
GHP_8 M2:  KIC_6 M1:
KIC_6 M1: 
- After incubation, place all samples in the tabletop centrifuge and spin at 13000 rcf for 1 minute
- Make new set of labeled 1.5 ml tubes
- Pipette all supernatant (200 ul) to each new tube
- Make 1.5 ml tube of 10 mM Tris HCl pH 8 and place in thermomixer at 70 deg C
- Set up tubes for extraction - 1 yellow spin column inside a collection tube for each sample - label lid of spin column
- Get liquid waste beaker from sink near -80 freezers
- Add 2 parts volume (400 ul) of Genomic Binding buffer to each tube
- Vortex for 5 seconds and spin down in mini centrifuge
- Add 400 ul of sample to their labeled yellow spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Put spin columns in same collection tubes
- Add remaining liquid (200 ul) from each sample to labeled yellow spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Transfer spin columns to new collection tubes and discard of old collection tubes
- Add 400 ul of DNA pre-wash buffer to each spin column
- Centrifuge at 13000 rcf from 1 minute
- Pour off flow through in liquid waste beaker
- Place spin columns in same collection tubes
- Add 700 ul of g-DNA wash buffer to each spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Put spin columns in same collection tubes
- Add 200 ul of g-DNA wash buffer to each spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Make final 1.5 ml tubes - label lide with sample id and DNA; label side with initials, date of extraction, sample id, DNA, and C. virginica
- Transfer spin columns to labeled 1.5 ml tubes
- Pour off flow through in liquid waste beaker and discard collection tubes
- Take warmed 10 mM Tris HCl pH 8 out of thermomixer
- Add 50 ul of warmed 10 mM Tris HCl pH 8 to each spin column by dripping directly over the filter without touching it
- Added too much Tris to KIC_6, so I’ll add ~25 ul of Tris during the second elution for this sample
- Incubate for 5 minutes
- Place tubes in centrifuge with all the lids of the 1.5 ml tubes facing the same direction and centrifuge at MAX speed for 1 minute
- Take tubes out - DO NOT pour off liquid, keep in tube
- Add an additional 50 ul of warmed 10 mM Tris HCl pH 8 to each spin column by dripping directly over filter without touching it
- 100 ul of DNA total
- Only 25 ul of Tris added to KIC_6 for this elution
- Incubate for 5 minutes
- Centrifuge at max speed for 1 minute
- Take out spin columns at discard
- Aliquot 6 ul of each sample to their respective PCR strip tube
- 3 ul for agarose gel
- 1 ul for Qubit
- 1 ul for TapeStation (if needed)
- 1 ul for error
94 ul remaining in 1.5 ml sample tubes
_Qubit dsDNA BR assay__
The captured pools were quantified following Qubit protocol for BR DNA
- 199 ul x 7.2 samples = 1432.8 ul of Buffer
- 1 ul x 7.2 samples = 7.2 ul of reagent
| Sample | Avg ng/μl |
|---|---|
| Std 1 | 179 RFU |
| Std 2 | 18232 RFU |
| NAR_7 | 39.6 |
| BAR_5 | 30.6 |
| BAR_7 | 50.2 |
| GHP_8 | 43.9 |
| KIC_6 | 17.2 |
Agarose Gel Electrophoresis
The DNA quality and size were assessed following Agarose Gel Protocol for a small 1% gel.
- Only 3 ul of DNA for each sample were used
- Used a diluted gelgreen made by Maggie Schedl
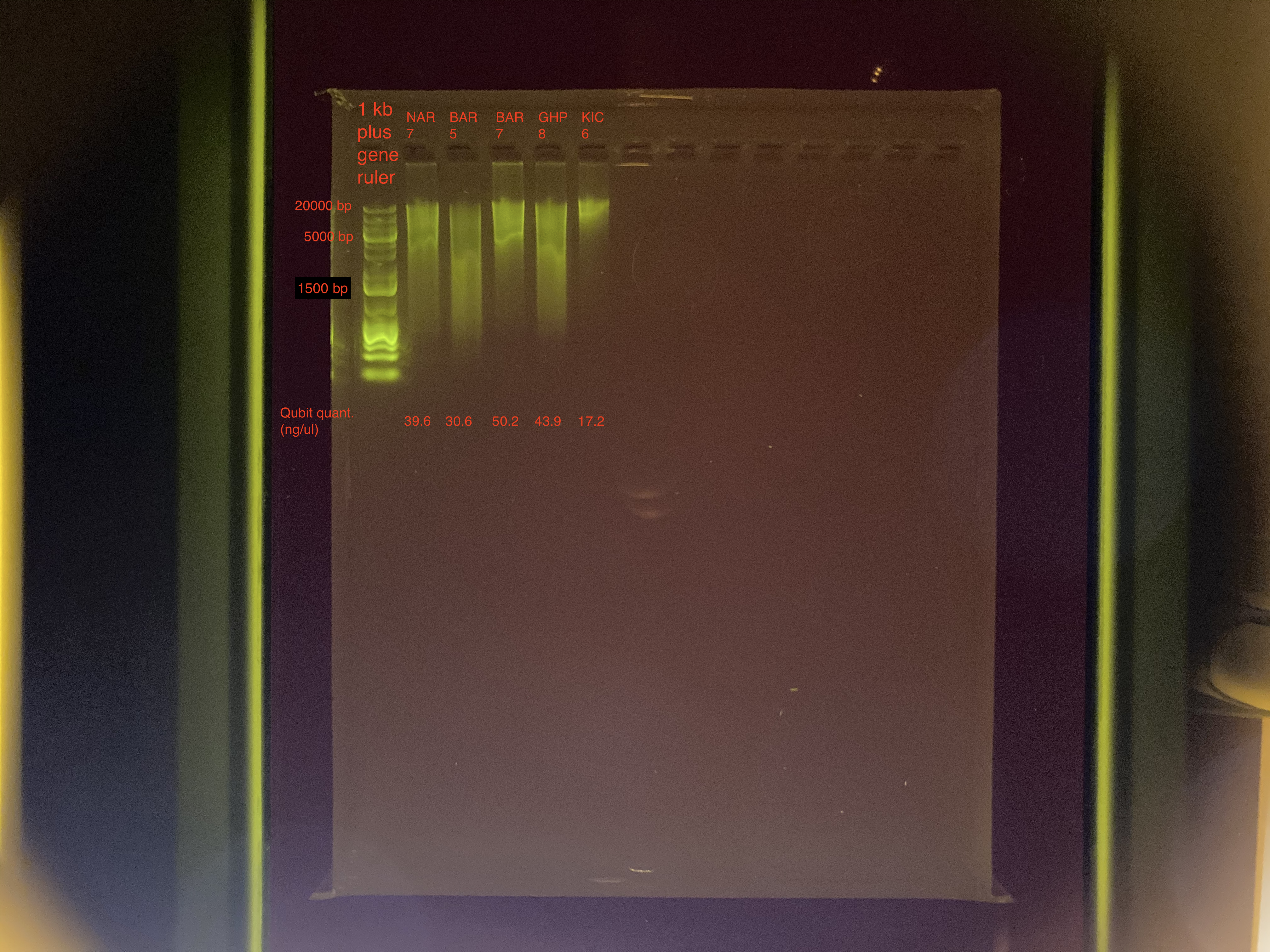
The samples still look a bit streaky, so I’ll run them on a TapeStation.
TapeStation
Completed on January 20, 2021
Samples were run on the TapeStation following the protocol for Genomic DNA.
- Ran samples BAR_5 and KIC_6
See full report here.
KIC_6 looked really good, BAR_5 had a lot of smaller fragments around 1500 bp. Next time I will try smaller pieces of tissue.
