Narragansett Bay Adult Oyster DNA Extractions Part 2
DNA Extraction of Adult Oyster Tissue - Part 2
~10 wild adult oysters were collected from 5 populations in Narragansett Bay
- Narrow River
- Green Hill Pond
- Barrington River
- Kickemuit
- Mary C. Donovan Marsh
Mantle and gill tissue were dissected from the oysters. Dissection protocol can be accessed here.
DNA Extractions
Completed on January 20, 2021
Zymo Research Quick-DNA Miniprep Plus used for DNA extractions of adult oyster tissue
- Pull samples out of -80 freezer and put on ice
- NAR_4
- GHP_2
- KIC_2
- MCD_1
- MCD_4
- Pull Proteinase k out of upright -20 freezer in Puritz Zymo Reagents box and put on ice
- Pull out Blue Solid Tissue Buffer from Zymo Research Quick DNA Miniprep kit
- First, I’m going to cut the tissue pieces into smaller pieces
- Make new labeled 1.5-ml tubes
- Lay out piece of aluminum foil
- Sterilize foil, razor blade, and forceps with 70% EtOH and Type II DI water
- Using forceps, take tissue piece out and lay on foil
- Use razor blade to cut small piece of tissue
- Transfer small tissue piece to new labeled 1.5-ml tube and return other tissue piece back to original tube
- Sterilize foil, razor blade, and forceps before each new sample
NAR_4:  GHP_2:
GHP_2:  KIC_2:
KIC_2:  MCD_1:
MCD_1:  MCD_4:
MCD_4: 
- In each tube with the small tissue piece, add 95 ul of nuclease-free water, 95 ul of solid tissue buffer, and 10 ul of proteinase k
- Vortex for 10 seconds and spin down on mini centrifuge
- Place tubes in thermomixer at 55 deg C shaking at 1200 rpm
- Check tissue after 45 minutes
- Added 5 more minutes
- After incubation, place all samples in the tabletop centrifuge and spin at 13000 rcf for 1 minute
- Make new set of labeled 1.5 ml tubes
- Pipette all supernatant (200 ul) to each new tube
- Make 1.5 ml tube of 10 mM Tris HCl pH 8 and place in thermomixer at 70 deg C
- Set up tubes for extraction - 1 yellow spin column inside a collection tube for each sample - label lid of spin column
- Get liquid waste beaker from sink near -80 freezers
- Add 2 parts volume (400 ul) of Genomic Binding buffer to each tube
- Vortex for 5 seconds and spin down in mini centrifuge
- Add 400 ul of sample to their labeled yellow spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Put spin columns in same collection tubes
- Add remaining liquid (200 ul) from each sample to labeled yellow spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Transfer spin columns to new collection tubes and discard of old collection tubes
- Add 400 ul of DNA pre-wash buffer to each spin column
- Centrifuge at 13000 rcf from 1 minute
- Pour off flow through in liquid waste beaker
- Place spin columns in same collection tubes
- Add 700 ul of g-DNA wash buffer to each spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Put spin columns in same collection tubes
- Add 200 ul of g-DNA wash buffer to each spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Make final 1.5 ml tubes - label lide with sample id and DNA; label side with initials, date of extraction, sample id, DNA, and C. virginica
- Transfer spin columns to labeled 1.5 ml tubes
- Pour off flow through in liquid waste beaker and discard collection tubes
- Take warmed 10 mM Tris HCl pH 8 out of thermomixer
- Add 50 ul of warmed 10 mM Tris HCl pH 8 to each spin column by dripping directly over the filter without touching it
- Incubate for 5 minutes
- Place tubes in centrifuge with all the lids of the 1.5 ml tubes facing the same direction and centrifuge at MAX speed for 1 minute
- Take tubes out - DO NOT pour off liquid, keep in tube
- Add an additional 50 ul of warmed 10 mM Tris HCl pH 8 to each spin column by dripping directly over filter without touching it
- Incubate for 5 minutes
- Centrifuge at max speed for 1 minute
- Take out spin columns at discard
- Aliquot 2 ul of each sample to their respective PCR strip tube for agarose gel
- 1 ul will be taken directly from 1.5 ml tube for Qubit
97 ul remaining in 1.5 ml sample tubes
Qubit dsDNA BR assay
The captured pools were quantified following Qubit protocol for BR DNA
- 199 ul x 7.2 samples = 1432.8 ul of Buffer
- 1 ul x 7.2 samples = 7.2 ul of reagent
| Sample | Avg ng/μl |
|---|---|
| Std 1 | 175 RFU |
| Std 2 | 16908 RFU |
| NAR_4 | 19.9 |
| GHP_2 | 33.6 |
| KIC_2 | 17.6 |
| MCD_1 | 2.48 |
| MCD_4 | 5.79 |
Agarose Gel Electrophoresis
The DNA quality and size were assessed following Agarose Gel Protocol for a small 1% gel.
- Only 2 ul of DNA for each sample were used
- Used a diluted gelgreen made by Maggie Schedl
- Used a different loading dye - Purple loading dye versus Tritrack
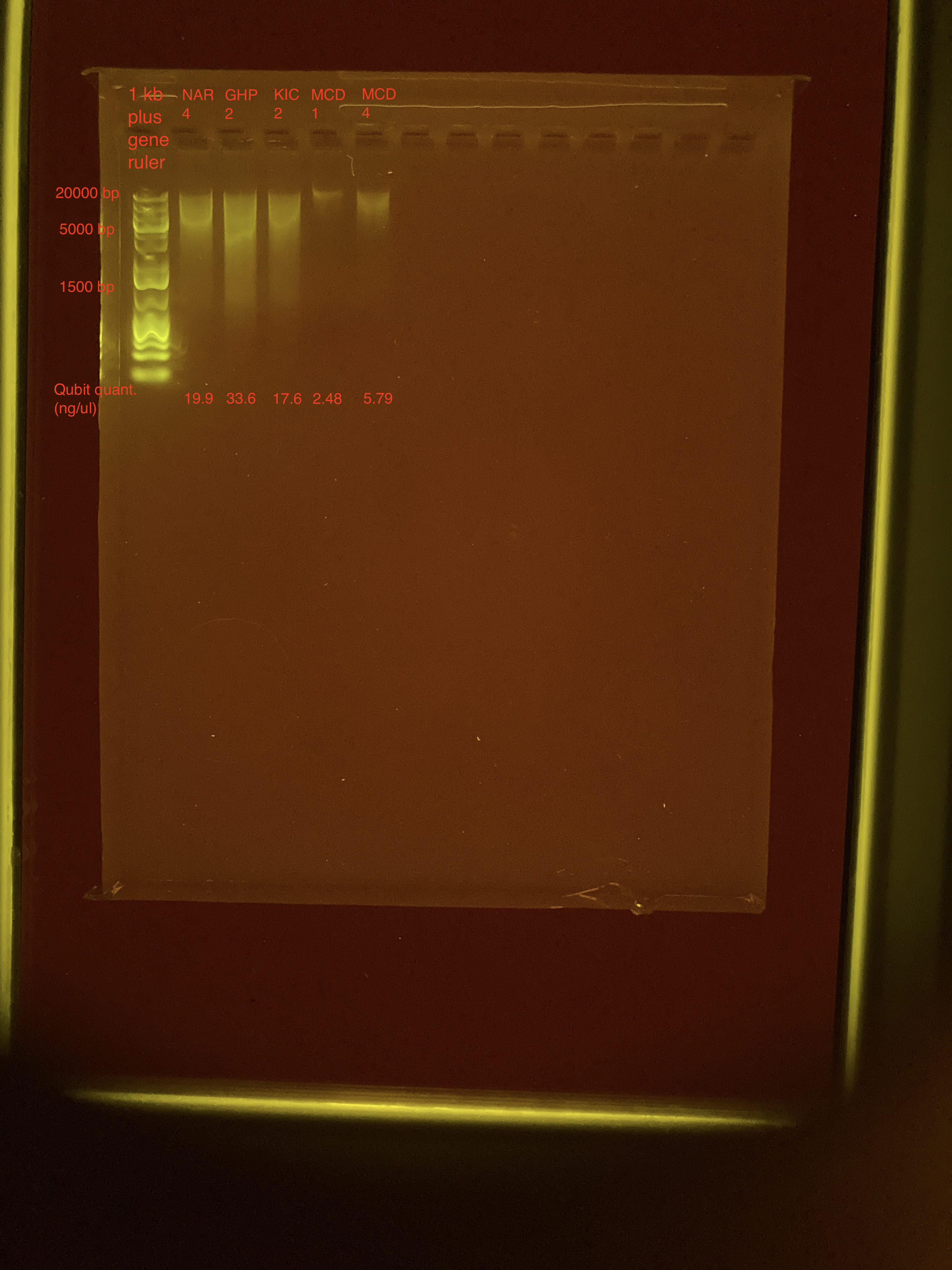
This gel looks better. The two MCD samples have very little DNA, so I will likely re-do the extraction using the full tissue pieces for these two. I think I’ll continue moving forward with the extractions using full tissue pieces.
Plan for next time:
- Try extraction on gill sample
- Do a PCB soak of the tissue before solubilization
- During solubilization in thermomixer, use slower rpm speed
- For centrifugation after solubilization, spin for 2 minutes at 13000 rcf
- During elution with 10 mM Tris HCl, separate the two elutions into two tubes
- Smaller DNA fragments will elute first
