Narragansett Bay Adult Oyster DNA Library Prep Part 1
DNA Library Prep for NB Adult Oyster EecSeq - 8 Samples
Initial bead cleanup and library prep were performed on Oct. 15, 2019.
Using the KAPA HyperPrep DNA Library Prep Kit on 8 DNA samples in 10 mM Tris HCl pH 8 containing 500 ng from the first 50 samples of the NB adult oyster experiment.
Previously, all samples were sonicated to 150 bp following the QSonica protocol- instructions can be found here.
A 1.8X bead cleanup was performed to concentrate 500 ng of DNA in 25 μl before library prep:
- Made fresh 80% EtOH
- Took KAPA Pure Beads out of fridge beforehand to warm to room temp
- Vortex and spin down DNA samples
- Added 90 μl (1.8 x 50 μl) of KAPA Pure Beads to each sample and pipette up and down 10 times to mix (avoid bubbles)
- Placed tubes on shaker at room temp for 15 minutes - shaker set to 200 rpm’s
- After 15 minute incubation, placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 25 μl of 10 mM Tris HCl pH 8 and incubated tubes on shaker at room temp for 5 minutes
- Placed tubes on magnet plate and transferred supernatant when clear to new labeled PCR strip tubes
End Repair and A-tailing
- Prepared end repair and a-tailing master mix:
- ERAT buffer: 3.5 μl * 9 = 31.5 μl
- ERAT enzyme: 1.5 μl * 9 = 13.5 μl
- In the 8 PCR strip tubes containing the 25 μl of 500 ng sheared DNA, added 5 μl of ERAT master mix
- Vortexed and spun down
- Placed samples in thermocycler A-tailing program in JONP login - program runs for ~ 1 hour
Adapter Ligation
Working adapter stocks 1-7, and 9-12 were diluted on Oct. 10, 2019; working adapter stock 8 was diluted on Oct. 21, 2019. Dilution was completed as followed:
- In new PCR strip tubes, added 20 μl of previously made annealed adapter stocks
-
Added 20 μl of Nuclease free water to annealed adapter stocks for 40 μl total of 20 μM adapter stocks
- Prepared adapter ligation master mix:
- Ligation buffer: 15 μl * 9.5 = 142.5 μl
- DNA ligase: 5 μl * 9.5 = 47.5 μl
- Nuclease free water: 2.5 μl * 9.5 = 23.75 μl
- Added 22.5 μl of ligation master mix and appropriately planned adapters to each sample. Adapters were added last to minimize adapter-adapter ligation.
| Sample | LMM | Adapter |
|---|---|---|
| 1.2 | 22.5μl | 2.5μl 2 |
| 1.6 | 22.5μl | 2.5μl 6 |
| 2.6 | 22.5μl | 2.5μl 4 |
| 3.9 | 22.5μl | 2.5μl 5 |
| 4.4 | 22.5μl | 2.5μl 10 |
| 4.7 | 22.5μl | 2.5μl 1 |
| 5.6 | 22.5μl | 2.5μl 10 |
| 5.8 | 22.5μl | 2.5μl 12 |
- Pipetted up and down to mix - pipette set to 50 μl
- spin down
- Incubated samples on shaker at room temp for 1 hour
0.8X Cleanup
- After incubation, added 44 μl of KAPA pure beads to each sample and pipetted up and down 10 times to mix (avoid bubbles)
- Placed tubes on shaker at room temp for 15 minutes
- Placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 12.5 μl 10 mM Tris HCl pH 8 and incubated tubes on shaker for 5 minutes
- Placed tubes back onto the magnet plate and removed supernatant when clear to new labeled PCR strip tubes
Library Amplification
- Every 5 samples get a different index primer pair for amplification. For these 8 samples, 7 different master mixes were made:
- Amp MM A - Sample 1.2
- 12.5 μl HotStart Ready mix
- 1.25 μl 501 primer
- 1.25 μl 701 primer
- Amp MM B - Sample 1.6
- 12.5 μl HotStart Ready mix
- 1.25 μl 502 primer
- 1.25 μl 702 primer
- Amp MM C - Sample 2.6
- 12.5 μl HotStart Ready mix
- 1.25 μl 504 primer
- 1.25 μl 704 primer
- Amp MM D - Sample 3.9
- 12.5 μl HotStart Ready mix
- 1.25 μl 506 primer
- 1.25 μl 706 primer
- Amp MM E - Sample 4.4
- 12.5 μl HotStart Ready mix
- 1.25 μl 507 primer
- 1.25 μl 707 primer
- Amp MM F - Sample 4.7
- 12.5 μl HotStart Ready mix
- 1.25 μl 508 primer
- 1.25 μl 708 primer
- Amp MM G - Samples 5.6 & 5.8
- 12.5 μl HotStart Ready mix * 2 = 25 μl
- 1.25 μl 510 primer * 2 = 2.5 μl
- 1.25 μl 710 primer * 2 = 2.5 μl
- Prepared new PCR tubes for the amplification with the following:
| Sample | volume adapter added DNA of sample | volume of Amp MM |
|---|---|---|
| 1.2 | 10μl | 15μl Amp MM A |
| 1.6 | 10μl | 15μl Amp MM B |
| 2.6 | 10μl | 15μl Amp MM C |
| 3.9 | 10μl | 15μl Amp MM D |
| 4.4 | 10μl | 15μl Amp MM E |
| 4.7 | 10μl | 15μl Amp MM F |
| 5.6 | 10μl | 15μl Amp MM G |
| 5.8 | 10μl | 15μl Amp MM G |
- Vortexed and spun down
- Placed samples in the thermocycler Genomic PCR program
1X Cleanup
- After PCR, added 25 μl of KAPA pure beads to each sample and pipetted up and down 10 times to mix (avoid bubbles)
- Placed tubes on shaker at room temp for 15 minutes
- Placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 16 μl 10 mM Tris HCl pH 8 and incubated tubes on shaker for 5 minutes
- Placed tubes back onto the magnet plate and removed supernatant when clear to new labeled PCR strip tubes
Qubit and TapeStation
Completed on Oct. 16, 2019
- Followed Qubit protocol for BR DNA
| Sample | Avg ng/μl |
|---|---|
| Std 1 | 191 RFU |
| Std 2 | 25292 RFU |
| 1.2 | 68.2 |
| 1.6 | 106.5 |
| 2.6 | 97.4 |
| 3.9 | 100 |
| 4.4 | 91.2 |
| 4.7 | 101 |
| 5.6 | 46.8 |
| 5.8 | 66.4 |
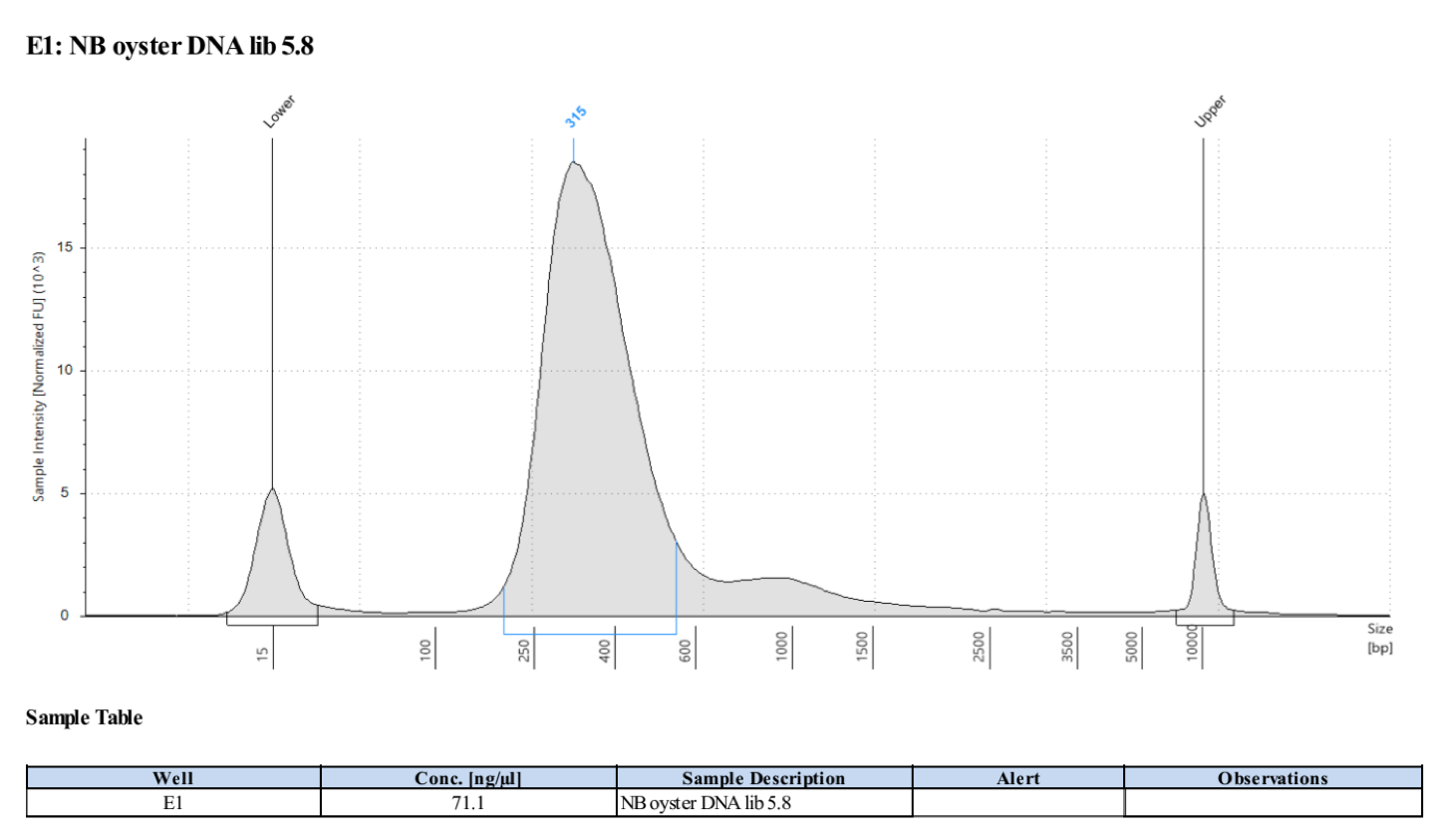
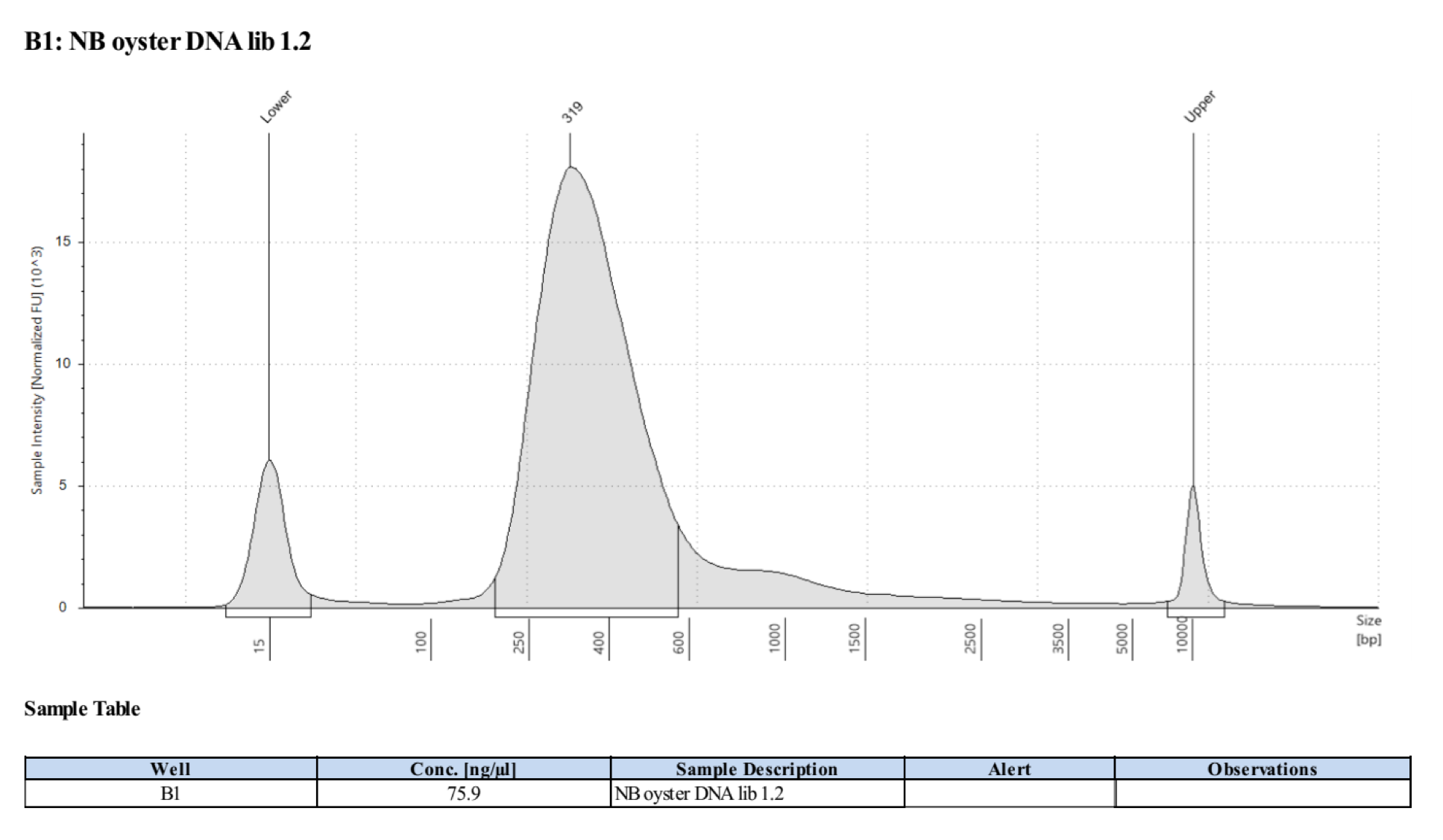
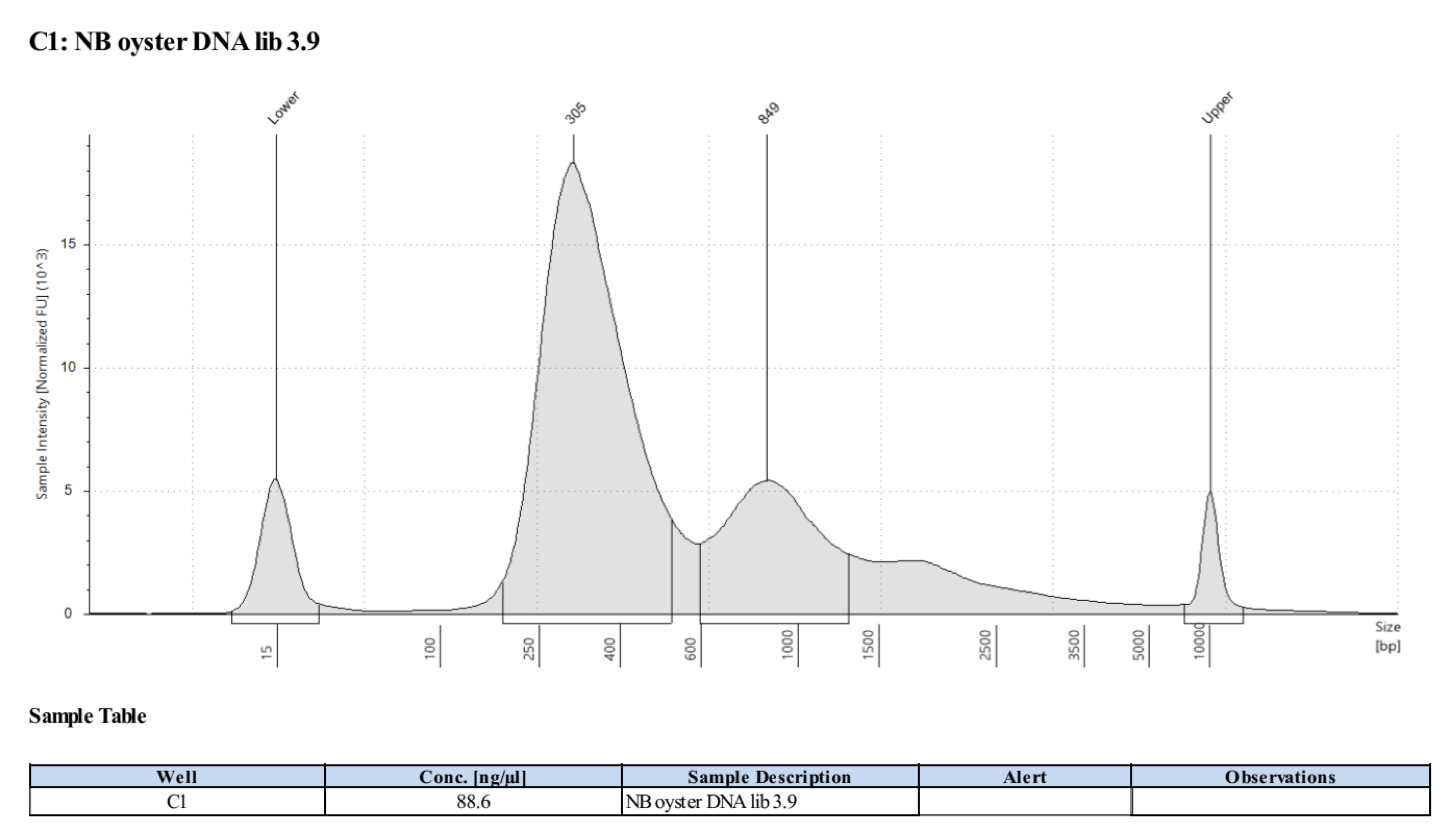
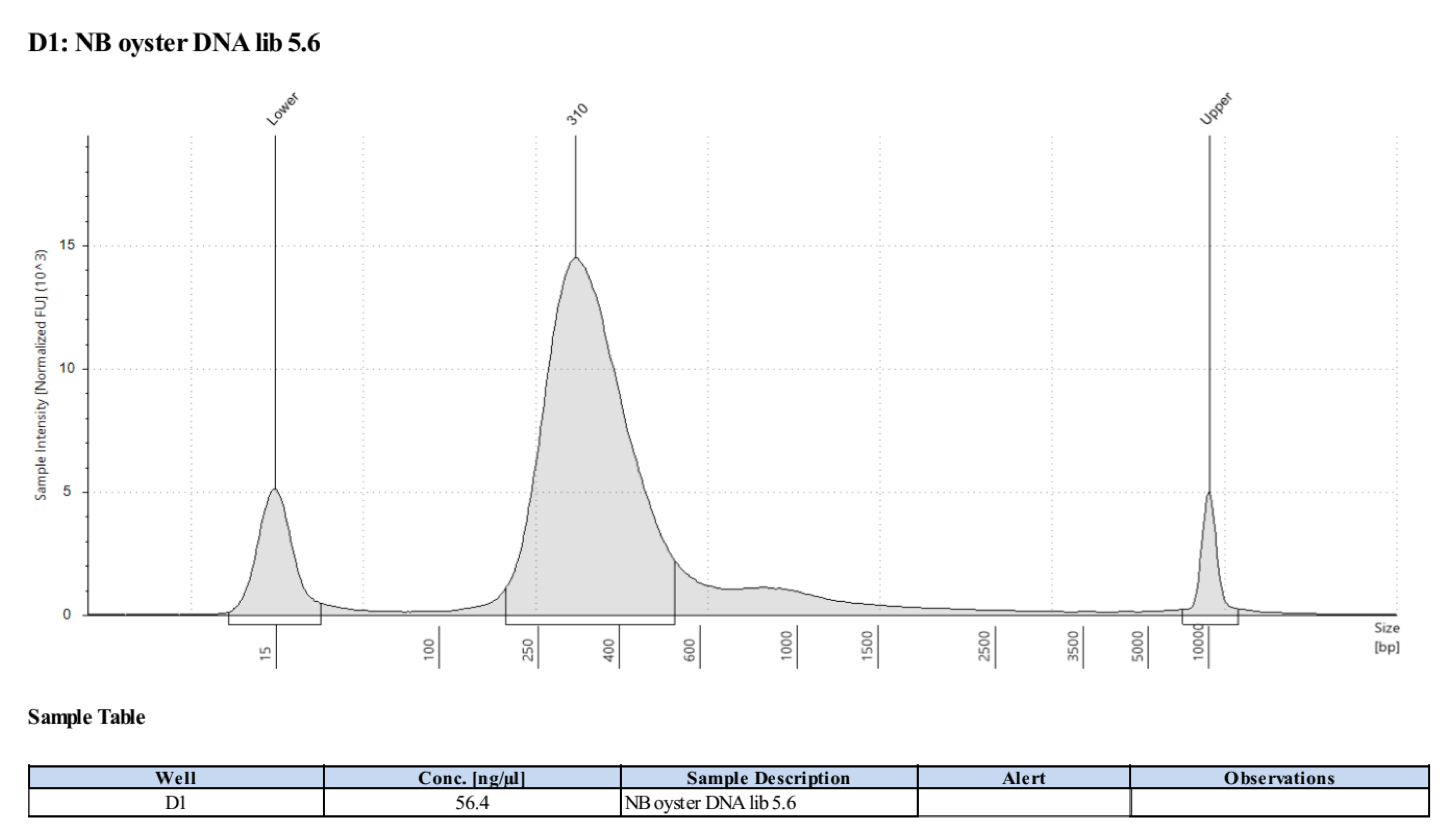
- Followed tapestation protocol for D5000 tapes on 4 representative samples to check
See full report here
Sample 1.2:
Sample 3.9:
Sample 5.6:
Sample 5.8: