Narragansett Bay Adult Oyster DNA Library Prep Part 3
DNA Library Prep for NB Adult Oyster EecSeq - 14 Samples
Initial bead cleanup was performed on Oct. 21, 2019 and library prep was performed on Oct. 30, 2019.
Using the KAPA HyperPrep DNA Library Prep Kit on 14 DNA samples in 10 mM Tris HCl pH 8 containing 500 ng from the first 50 samples of the NB adult oyster experiment.
Previously, all samples were sonicated to 150 bp following the QSonica protocol- instructions can be found here.
A 1.8X bead cleanup was performed to concentrate 500 ng of DNA in 25 μl before library prep:
- Made fresh 80% EtOH
- Took KAPA Pure Beads out of fridge beforehand to warm to room temp
- Vortex and spin down DNA samples
- Added 90 μl (1.8 x 50 μl) of KAPA Pure Beads to each sample and pipette up and down 10 times to mix (avoid bubbles)
- Placed tubes on shaker at room temp for 15 minutes - shaker set to 200 rpm’s
- After 15 minute incubation, placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 25 μl of 10 mM Tris HCl pH 8 and incubated tubes on shaker at room temp for 5 minutes
- Placed tubes on magnet plate and transferred supernatant when clear to new labeled PCR strip tubes
End Repair and A-tailing
- Prepared end repair and a-tailing master mix:
- ERAT buffer: 3.5 μl * 15 = 52.5 μl
- ERAT enzyme: 1.5 μl * 15 = 22.5 μl
- In the 14 PCR strip tubes containing the 25 μl of 500 ng sheared DNA, added 5 μl of ERAT master mix
- Vortexed and spun down
- Placed samples in thermocycler A-tailing program in JONP login - program runs for ~ 1 hour
Adapter Ligation
Working adapter stocks 1-7, and 9-12 were diluted on Oct. 10, 2019; working adapter stock 8 was diluted on Oct. 21, 2019. Dilution was completed as followed:
- In new PCR strip tubes, added 20 μl of previously made annealed adapter stocks
-
Added 20 μl of Nuclease free water to annealed adapter stocks for 40 μl total of 20 μM adapter stocks
- Prepared adapter ligation master mix:
- Ligation buffer: 15 μl * 15.5 = 232.5 μl
- DNA ligase: 5 μl * 15.5 = 77.5 μl
- Nuclease free water: 2.5 μl * 15.5 = 38.75 μl
- Added 22.5 μl of ligation master mix and appropriately planned adapters to each sample. Adapters were added last to minimize adapter-adapter ligation.
| Sample | LMM | Adapter |
|---|---|---|
| 1.1 | 22.5μl | 2.5μl 1 |
| 1.8 | 22.5μl | 2.5μl 8 |
| 1.10 | 22.5μl | 2.5μl 10 |
| 2.2 | 22.5μl | 2.5μl 12 |
| 2.5 | 22.5μl | 2.5μl 3 |
| 2.8 | 22.5μl | 2.5μl 6 |
| 3.2 | 22.5μl | 2.5μl 10 |
| 3.5 | 22.5μl | 2.5μl 1 |
| 3.8 | 22.5μl | 2.5μl 4 |
| 4.2 | 22.5μl | 2.5μl 8 |
| 4.5 | 22.5μl | 2.5μl 11 |
| 4.9 | 22.5μl | 2.5μl 3 |
| 5.2 | 22.5μl | 2.5μl 6 |
| 5.5 | 22.5μl | 2.5μl 9 |
- Pipetted up and down with multichannel to mix - pipette set to 50 μl
- spin down
- Incubated samples on shaker at room temp for 1 hour
0.8X Cleanup
- Made fresh 80% EtOH
- Took KAPA Pure Beads out of fridge beforehand to warm to room temp
- After incubation, added 44 μl of KAPA pure beads to each sample and pipetted up and down 10 times to mix (avoid bubbles)
- Placed tubes on shaker at room temp for 15 minutes
- Placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 12.5 μl 10 mM Tris HCl pH 8 and incubated tubes on shaker for 5 minutes
- Placed tubes back onto the magnet plate and removed supernatant when clear to new labeled PCR strip tubes
Library Amplification
- Every 5 samples get a different index primer pair for amplification. For these 14 samples, 9 different master mixes were made:
- Amp MM A - Sample 1.1
- 12.5 μl HotStart Ready mix
- 1.25 μl 501 primer
- 1.25 μl 701 primer
- Amp MM B - Samples 1.8 & 1.10
- 12.5 μl HotStart Ready mix * 2.2 = 27.5 μl
- 1.25 μl 502 primer * 2.2 = 2.75 μl
- 1.25 μl 702 primer * 2.2 = 2.75 μl
- Amp MM C - Samples 2.2 & 2.5
- 12.5 μl HotStart Ready mix * 2.2 = 27.5 μl
- 1.25 μl 503 primer * 2.2 = 2.75 μl
- 1.25 μl 703 primer * 2.2 = 2.75 μl
- Amp MM D - Sample 2.8
- 12.5 μl HotStart Ready mix
- 1.25 μl 504 primer
- 1.25 μl 704 primer
- Amp MM E - Samples 3.2 & 3.5
- 12.5 μl HotStart Ready mix * 2.2 = 27.5 μl
- 1.25 μl 505 primer * 2.2 = 2.75 μl
- 1.25 μl 705 primer * 2.2 = 2.75 μl
- Amp MM F - Sample 3.8
- 12.5 μl HotStart Ready mix
- 1.25 μl 506 primer
- 1.25 μl 706 primer
- Amp MM G - Samples 4.2 & 4.5
- 12.5 μl HotStart Ready mix * 2.2 = 27.5 μl
- 1.25 μl 507 primer * 2.2 = 2.75 μl
- 1.25 μl 707 primer * 2.2 = 2.75 μl
- Amp MM H - Sample 4.9
- 12.5 μl HotStart Ready mix
- 1.25 μl 508 primer
- 1.25 μl 708 primer
- Amp MM I - Samples 5.2 & 5.5
- 12.5 μl HotStart Ready mix * 2.2 = 27.5 μl
- 1.25 μl 509 primer * 2.2 = 2.75 μl
- 1.25 μl 709 primer * 2.2 = 2.75 μl
- Prepared new PCR tubes for the amplification with the following:
| Sample | volume adapter added DNA of sample | volume of Amp MM |
|---|---|---|
| 1.1 | 10μl | 15μl Amp MM A |
| 1.8 | 10μl | 15μl Amp MM B |
| 1.10 | 10μl | 15μl Amp MM B |
| 2.2 | 10μl | 15μl Amp MM C |
| 2.5 | 10μl | 15μl Amp MM C |
| 2.8 | 10μl | 15μl Amp MM D |
| 3.2 | 10μl | 15μl Amp MM E |
| 3.5 | 10μl | 15μl Amp MM E |
| 3.8 | 10μl | 15μl Amp MM F |
| 4.2 | 10μl | 15μl Amp MM G |
| 4.5 | 10μl | 15μl Amp MM G |
| 4.9 | 10μl | 15μl Amp MM H |
| 5.2 | 10μl | 15μl Amp MM I |
| 5.5 | 10μl | 15μl Amp MM I |
- Vortexed and spun down
- Placed samples in the thermocycler Genomic PCR program
1X Cleanup
- After PCR, added 25 μl of KAPA pure beads to each sample and pipetted up and down 10 times to mix (avoid bubbles)
- Placed tubes on shaker at room temp for 15 minutes
- Placed tubes on magnet plate and removed supernatant from tubes when it was fully clear not disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed supernatant from each tube on the magnet plate without disturbing the beads
- Added 200 μl of 80% EtOH to each tube while still the magnet not disturbing the beads
- Removed ALL the supernatant from each tube on the magnet plate without disturbing the beads. Extra EtOH blobs were removed with p20 pipette tips
- Resuspended beads in 16 μl 10 mM Tris HCl pH 8 and incubated tubes on shaker for 5 minutes
- Placed tubes back onto the magnet plate and removed supernatant when clear to new labeled PCR strip tubes
Qubit and TapeStation
Completed on Oct. 31, 2019
- Followed Qubit protocol for BR DNA
| Sample | Avg ng/μl |
|---|---|
| Std 1 | 173 RFU |
| Std 2 | 18987 RFU |
| 1.1 | 115 |
| 1.8 | 151 |
| 1.10 | 131.5 |
| 2.2 | 149.5 |
| 2.5 | 144 |
| 2.8 | 82.5 |
| 3.2 | 130 |
| 3.5 | 125 |
| 3.8 | 98.5 |
| 4.2 | 157.5 |
| 4.5 | 122.5 |
| 4.9 | 145.5 |
| 5.2 | 146.5 |
| 5.5 | 147.5 |
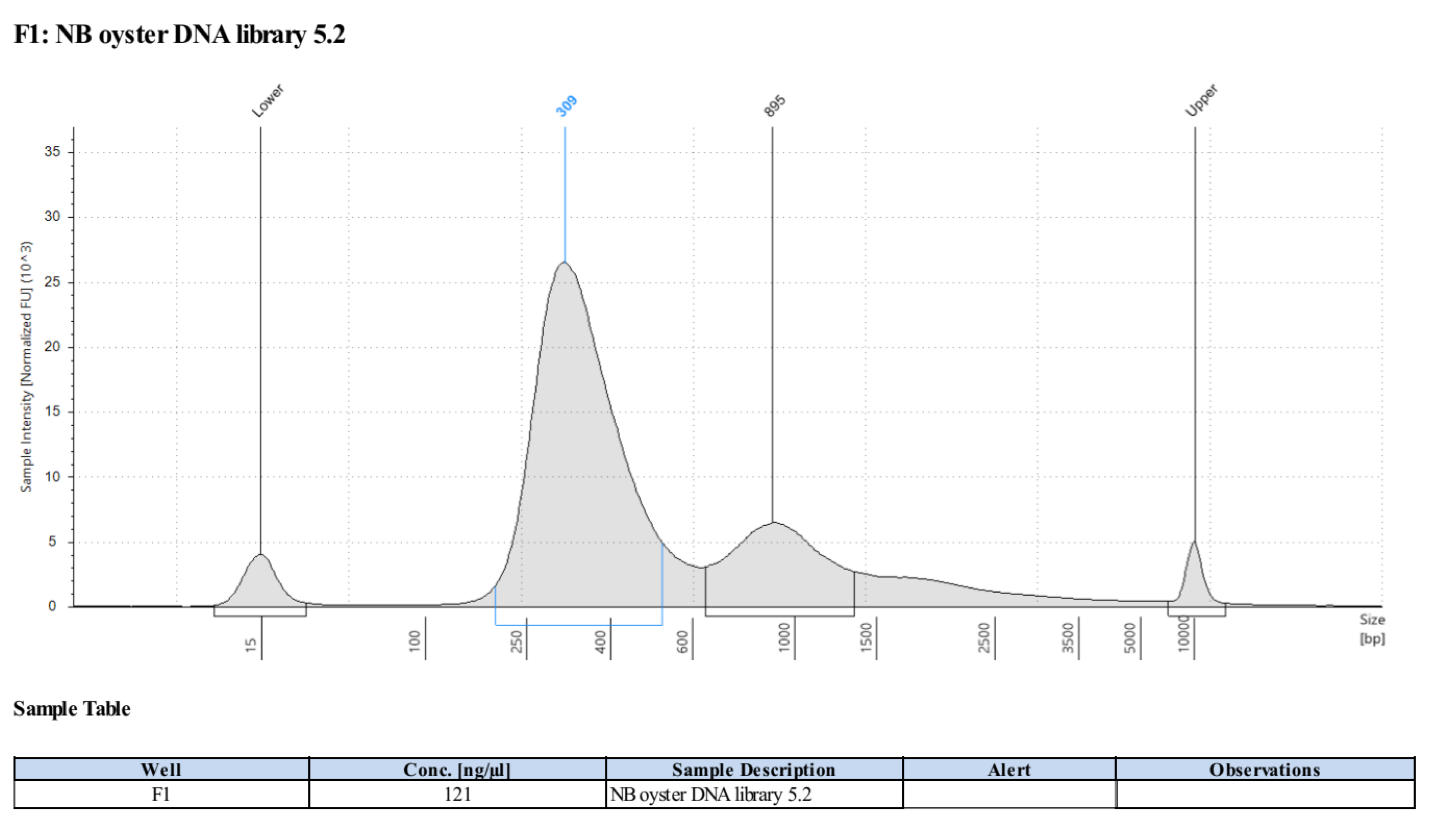
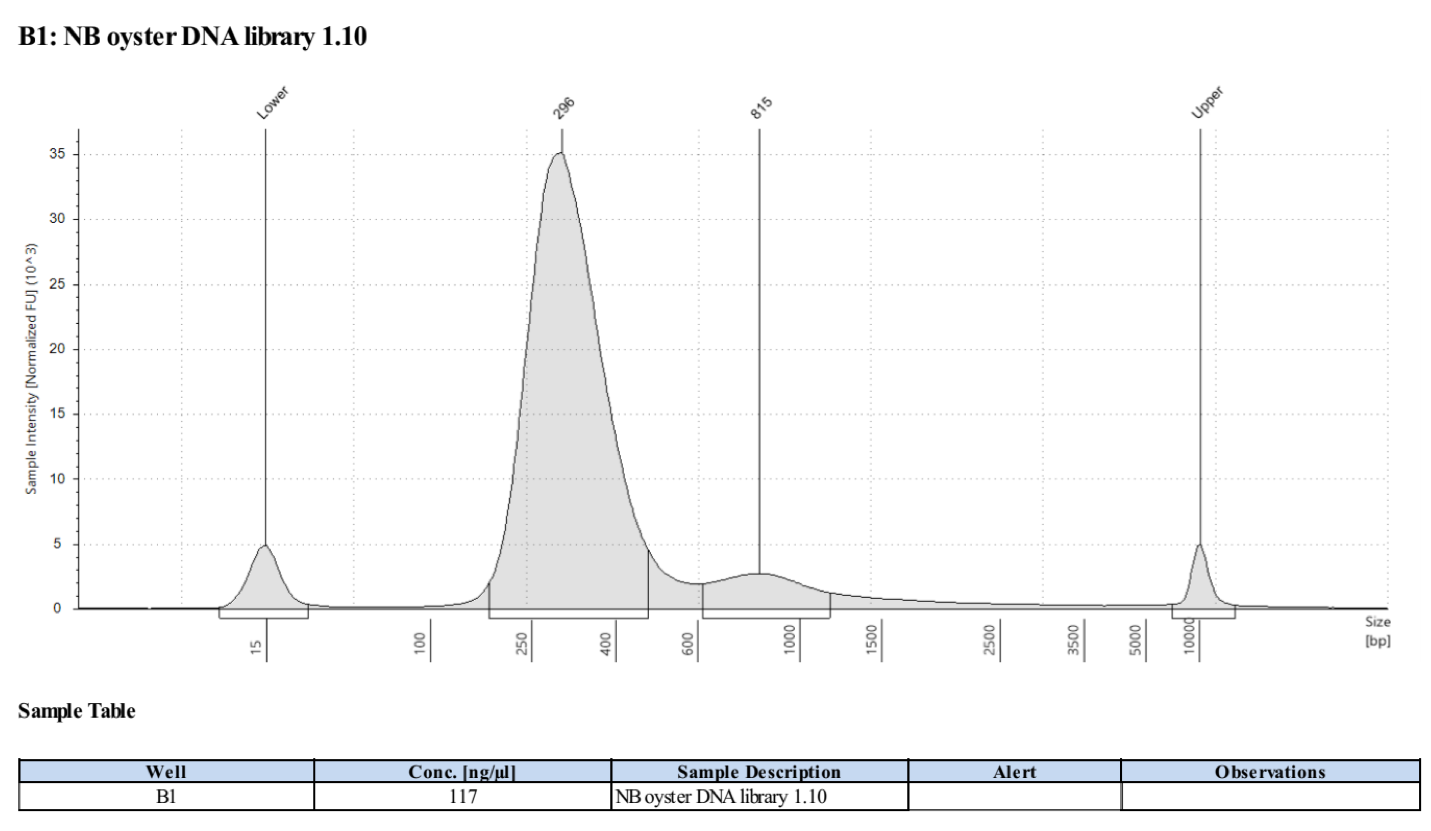
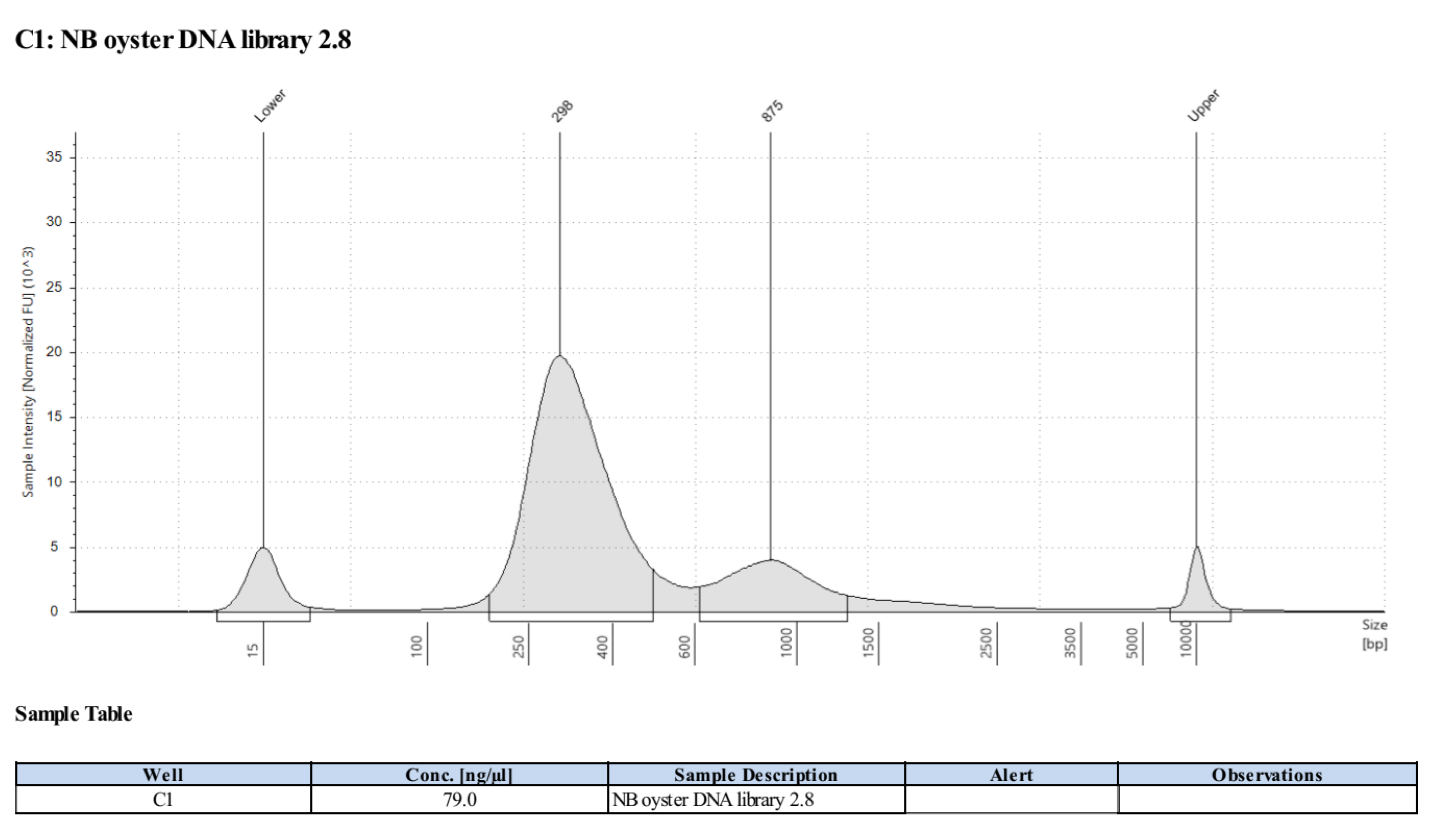
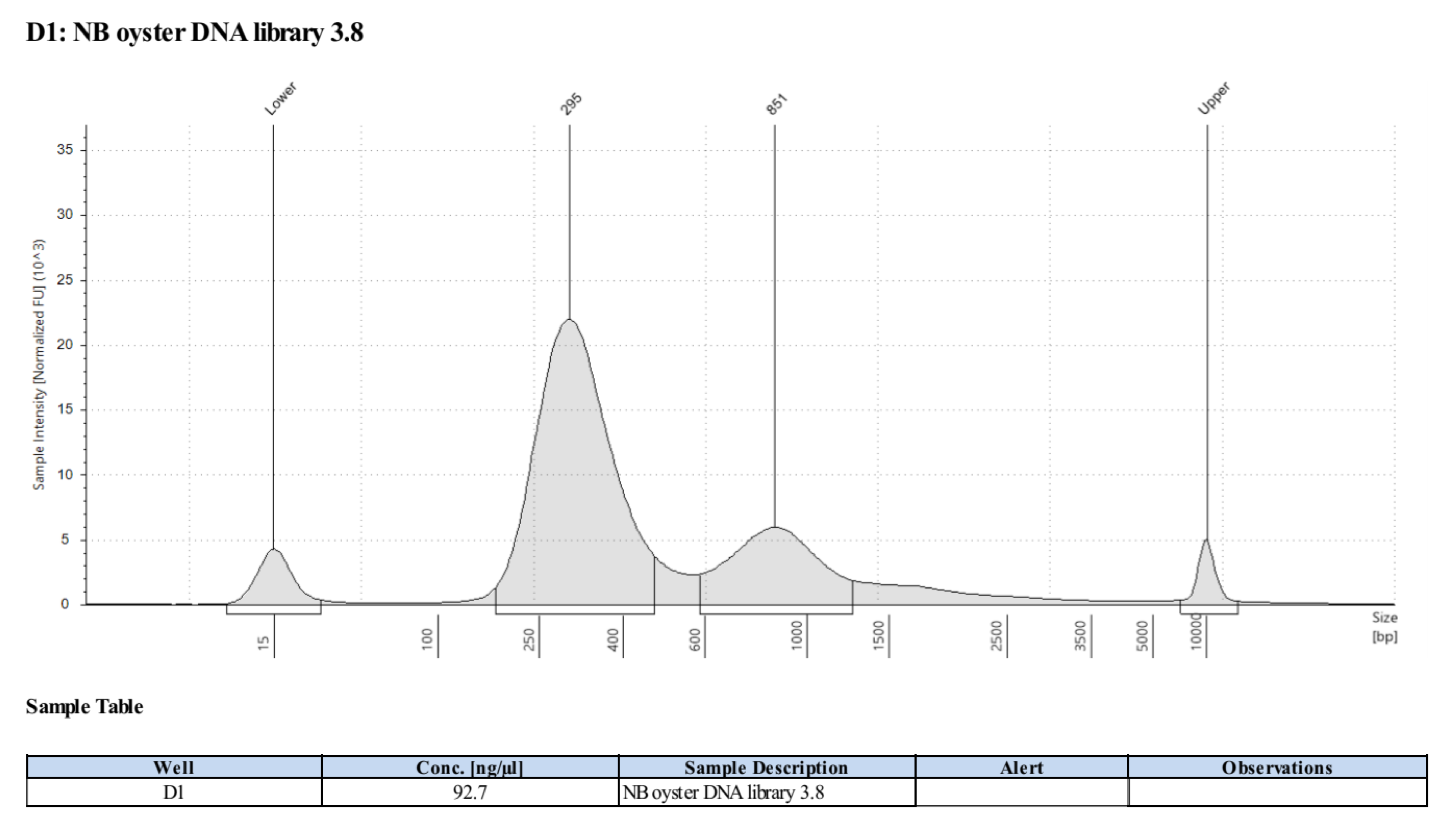
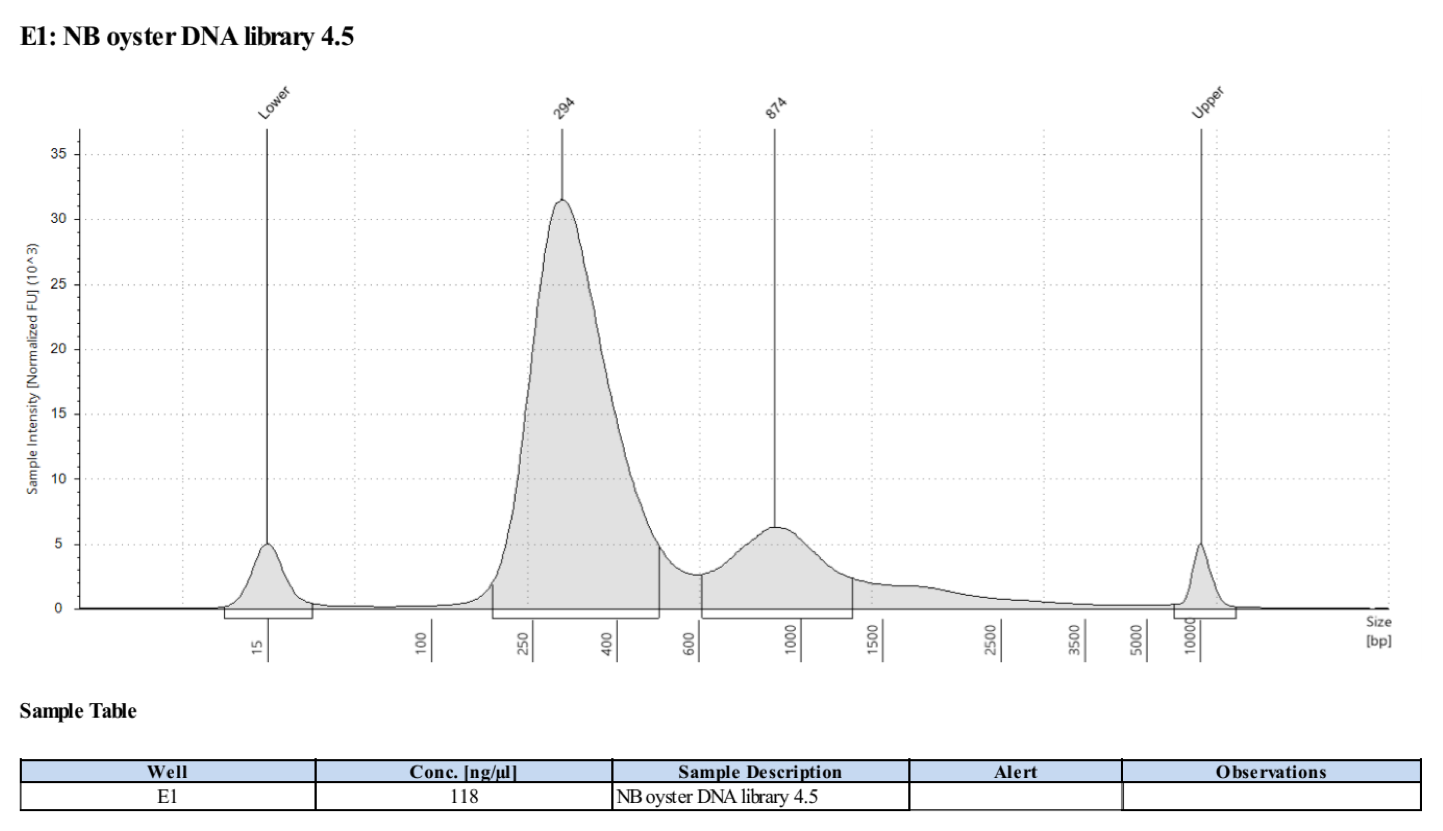
- Followed tapestation protocol for D5000 tapes on 5 representative samples to check
See full report here
Sample 1.10:
Sample 2.8:
Sample 3.8:
Sample 4.5:
Sample 5.2: