Narragansett Bay Adult Oyster Practice DNA Extractions
Practice DNA Extractions on Oysters from Fish Market
Before starting DNA extractions of the tissue from oysters collected at sites in Narragansett Bay, I wanted to practice the extraction protocol using the tissue of oysters purchased from the Fearless Fish Market. Mantle and gill tissues were dissected from these oysters in September 2020 by Jacob Green. Tissue dissection protocol can be accessed here.
DNA Extractions
Completed on November 11, 2020
Zymo Research Quick-DNA Miniprep Plus used for DNA extractions of adult oyster tissue
- Pull samples out of -80 and put on ice
- AA001 M1
- AA004 M1
- AA008 M1
- Pull Proteinase k out of upright -20 freezer in Puritz Zymo Reagents box and put on ice
- Pull out Blue Solid Tissue Buffer from Zymo Research Quick-DNA Miniprep Plus kit
- To each original tube containing tissue, add 95 ul of nuclease-free water, 95 ul of solid tissue buffer, and 10 ul of proteinase k
- Vortex for 10 seconds and spin down in mini centrifuge
- Place tubes in thermomixer at 55 deg C, shaking at 1200 rpm
- check the tissue after 1 hour of incubation
- Tissue was completely solubilized after 1 hour
AA001 Lid: 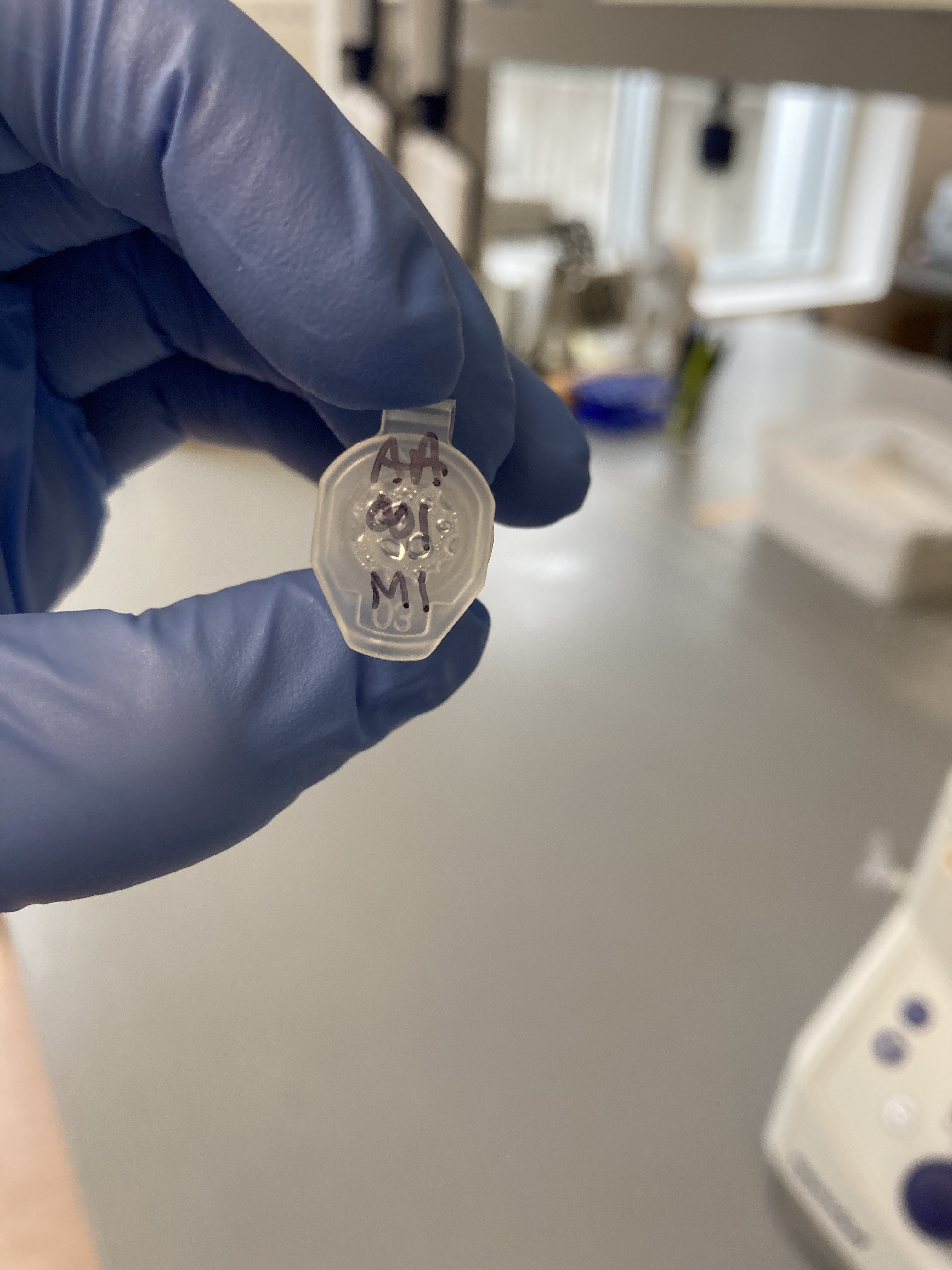 AA001 Side:
AA001 Side:  AA004 Lid:
AA004 Lid:  AA004 Side:
AA004 Side:  AA008 Lid:
AA008 Lid:  AA008 Side:
AA008 Side: 
- After incubation, place all samples in tabletop centrifuge at 13000 rcf for 1 minute
- Little debris pellet should form at bottom of tube
- Make new set of labeled 1.5-ml tubes
- Pipette all supernatant (200 ul) to new tube
- Make 1.5-ml tube of 10 mM Tris HCl pH 8 and place in thermomixer at 70 deg C
- Set up tubes for extraction - 1 yellow spin column inside a collection tube for each sample - label lid of spin column
- Get liquid waste beaker from sink near -80 freezers
- Add 2 parts volume (400 ul) of Genomic Binding buffer to each tube
- Vortex for 5 seconds and spin down in mini centrifuge
- Add 400 ul of sample to their labeled yellow spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Put spin columns in same collection tubes
- Add remaining liquid (200 ul) from each sample to labeled yellow spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Transfer spin columns to new collection tubes and discard of old collection tubes
- Add 400 ul of DNA pre-wash buffer to each spin column
- Centrifuge at 13000 rcf from 1 minute
- Pour off flow through in liquid waste beaker
- Place spin columns in same collection tubes
- Add 700 ul of g-DNA wash buffer to each spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Pour off flow through in liquid waste beaker
- Put spin columns in same collection tubes
- Add 200 ul of g-DNA wash buffer to each spin column
- Centrifuge spin columns at 13000 rcf for 1 minute
- Make final 1.5 ml tubes - label lide with sample id and DNA; label side with initials, date of extraction, sample id, DNA, and C. virginica
- Transfer spin columns to labeled 1.5 ml tubes
- Pour off flow through in liquid waste beaker and discard collection tubes
- Take warmed 10 mM Tris HCl pH 8 out of thermomixer
- Add 50 ul of warmed 10 mM Tris HCl pH 8 to each spin column by dripping directly over the filter without touching it
- Incubate for 5 minutes
- Place tubes in centrifuge with all the lids of the 1.5 ml tubes facing the same direction and centrifuge at MAX speed for 1 minute
- Take tubes out - DO NOT pour off liquid, keep in tube
- Add an additional 50 ul of warmed 10 mM Tris HCl pH 8 to each spin column by dripping directly over filter without touching it
- 100 ul of DNA total
- Incubate for 5 minutes
- Centrifuge at max speed for 1 minute
- Take out spin columns at discard
- Set up PCR strip tubes, labeling with the appropriate sample ids
- Aliquot 8 ul of each sample to their respective PCR strip tube
- 5 ul for agarose gel
- 1 ul for Qubit
- 1 ul for TapeStation (if needed)
- 1 ul for error
92 ul remaining in 1.5 ml sample tubes
Qubit dsDNA BR assay
The captured pools were quantified following Qubit protocol for BR DNA
- 199 ul x 5.2 samples = 1034.8 ul of Buffer
- 1 ul x 5.2 samples = 5.2 ul of reagent
| Sample | Avg ng/μl |
|---|---|
| Std 1 | 177 RFU |
| Std 2 | 17776 RFU |
| AA001 | 40.4 |
| AA004 | 66.5 |
| AA008 | 51 |
Agarose Gel Electrophoresis
The DNA quality and size were assessed following Agarose Gel Protocol for a small 1% gel.
Gel was run with Kevin Wong. His samples are the top row.
Gel_Practice_Part1: 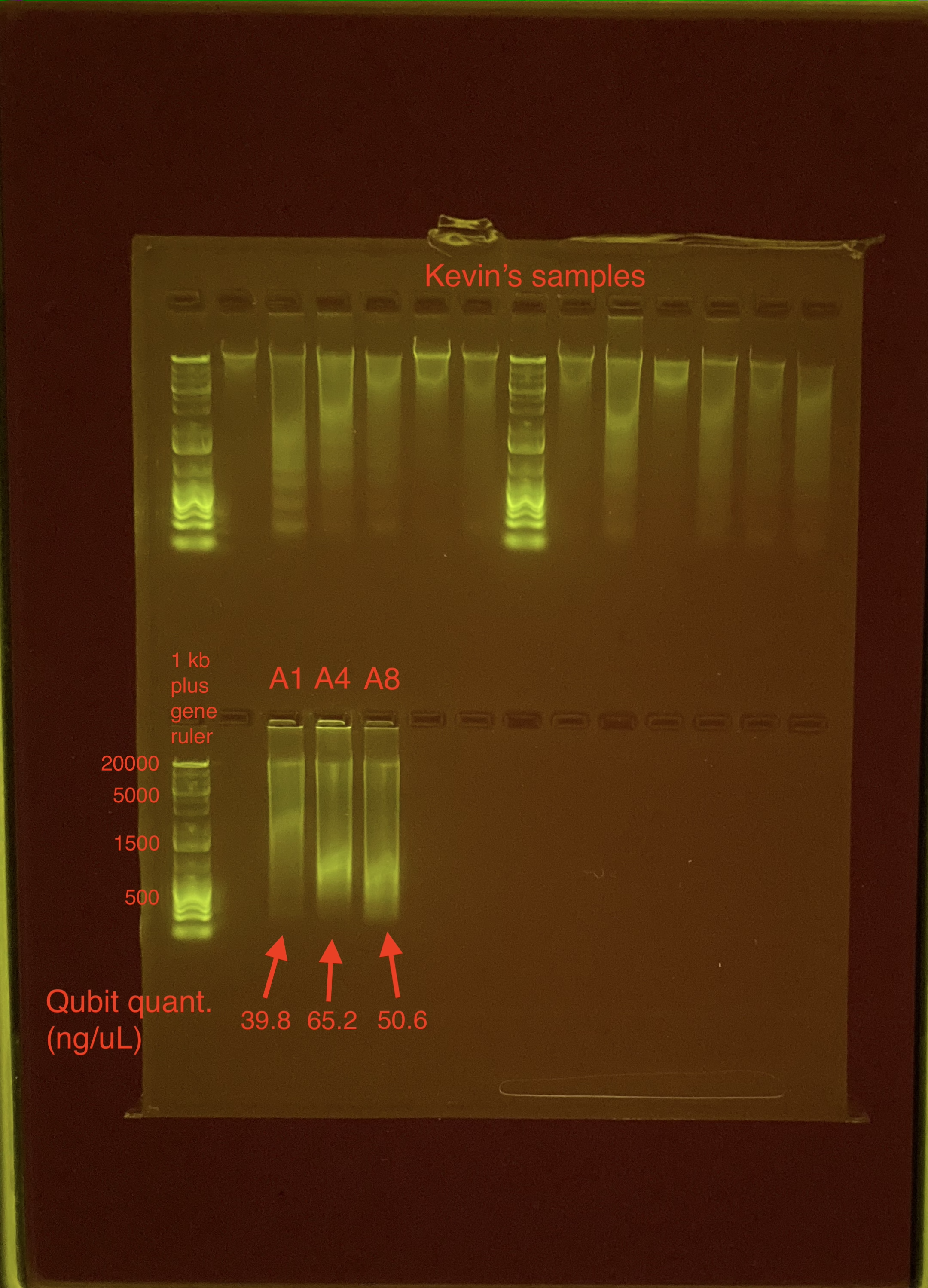
The samples look very streaky. Likely due to gel artifacts Will do a 1X Beadclean and then run samples on gel again.
1X Beadclean
Completed on November 18, 2020
- Pull samples out of freezer and put on ice
- Pull KAPA Pure Beads out of fridge and let warm to room temperature for ~30 minutes
- Make new batch of 80% EtOH
- Aliqout 25 ul of the DNA sample into new labeled PRCR strip tubes
- *67 ul saved in original 1.5-ml tubes in upright -20 freezer
- Add 25 ul of KAPA Pure Beads to each DNA sample and pipette to mix
- Incubate at room temperature on shaker for 15 minutes
- Place strip tubes on magnet, remove supernatant when clear
- dispose of in liquid waster beaker
- Add 200 ul of 80% EtOH while samples are still on magnet
- Remove clear supernatant
- Again, add 200 ul of 80% EtOH while still on magnet
- Remove all of clear supernatant
- Resuspend in 25 ul of 10mM Tris HCl pH 8 and incubate at room temperature for 5 minutes
- Put tubes on magent, transfer clear supernatant to new labeled tubes
- Transfer 5 ul of each sample to new labeled PCR strip tubes for agarose gel
Qubit dsDNA BR assay
The captured pools were quantified following Qubit protocol for BR DNA
| Sample | Avg ng/μl |
|---|---|
| Std 1 | 183 RFU |
| Std 2 | 19459 RFU |
| AA001 | 23.6 |
| AA004 | 60.5 |
| AA008 | 46.8 |
Agarose Gel Electrophoresis
The DNA quality and size were assessed following Agarose Gel Protocol for a small 1% gel.
Gel_Practice_Part2: 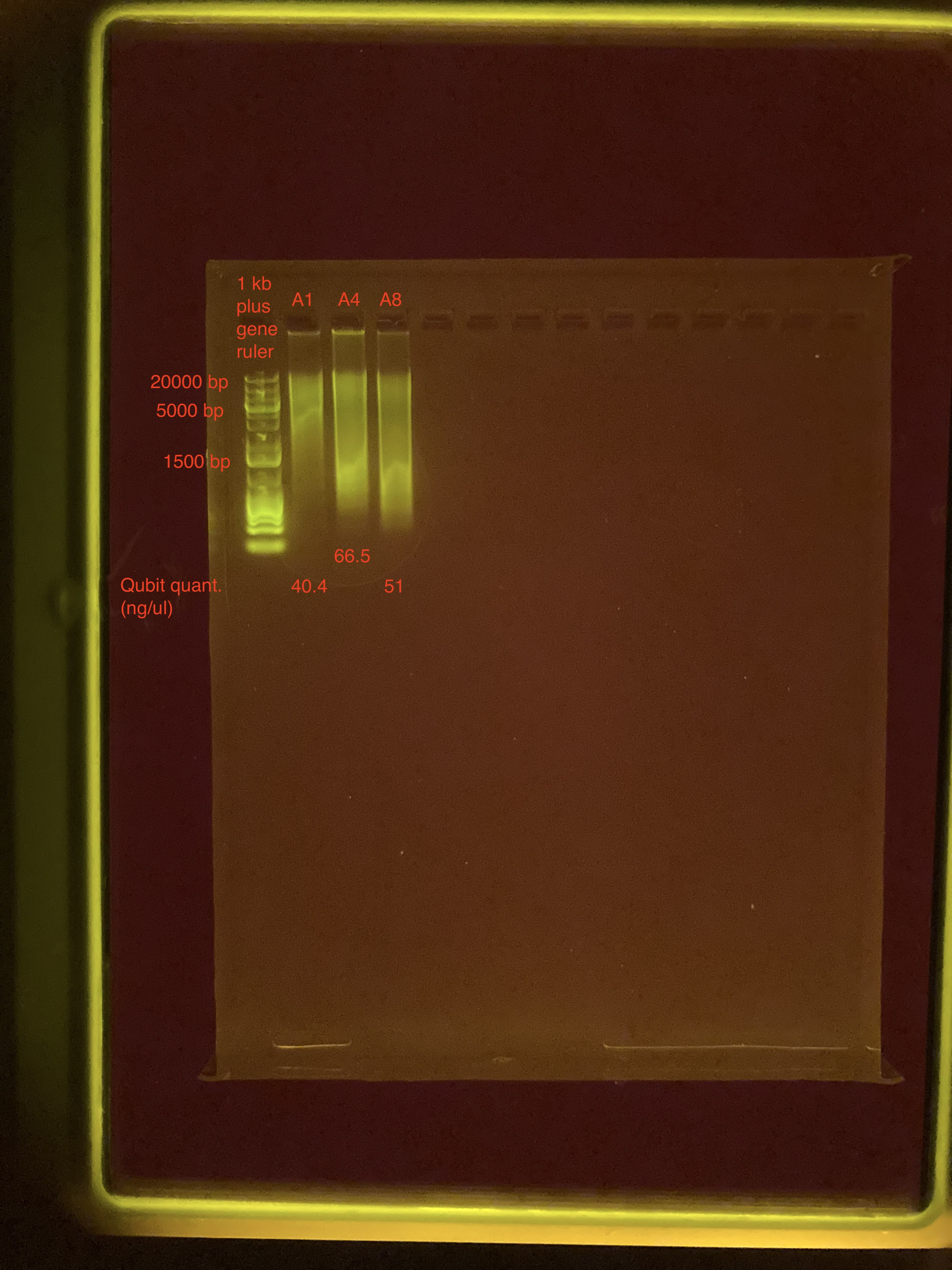
The gel does not look any different. I will look at the samples on the TapeStation.
TapeStation
Completed on November 25, 2020
Samples were run on the TapeStation following the protocol for Genomic DNA.
See full report here
The DNA looks pretty good, some smaller fragments around 1500 bp, but the majority is still larger fragments. I’m not sure why the gel looked bad. For future gels, I will use less DNA (2 ul instead of 5 ul) and used diluted gelgreen.
